environmental DNA
monitoring biodiversity across Maryland
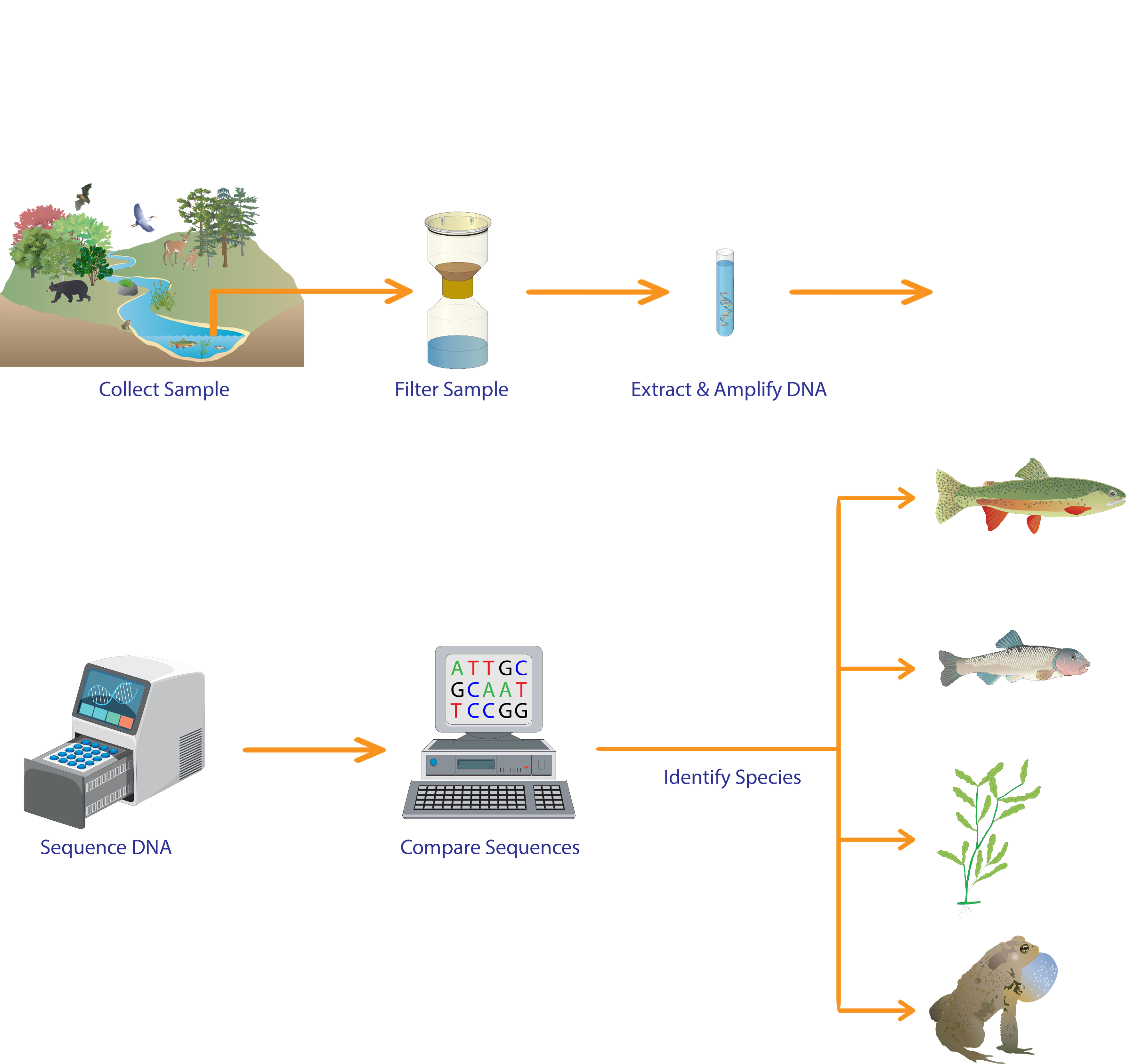
Environmental DNA (eDNA) allows us to detect organisms simply by sampling the water they live in. Just like humans shed skin cells and hair, aquatic organisms leave behind genetic traces in rivers and streams.
By collecting and analyzing water samples, we can identify the presence of fish, amphibians, invertebrates, and even invasive species—without ever seeing or capturing them.
eDNA is a powerful complement to traditional field surveys and can enhance biodiversity monitoring efforts across large spatial scales.
eDNA is a powerful complement to traditional field surveys and can enhance biodiversity monitoring efforts across large spatial scales.
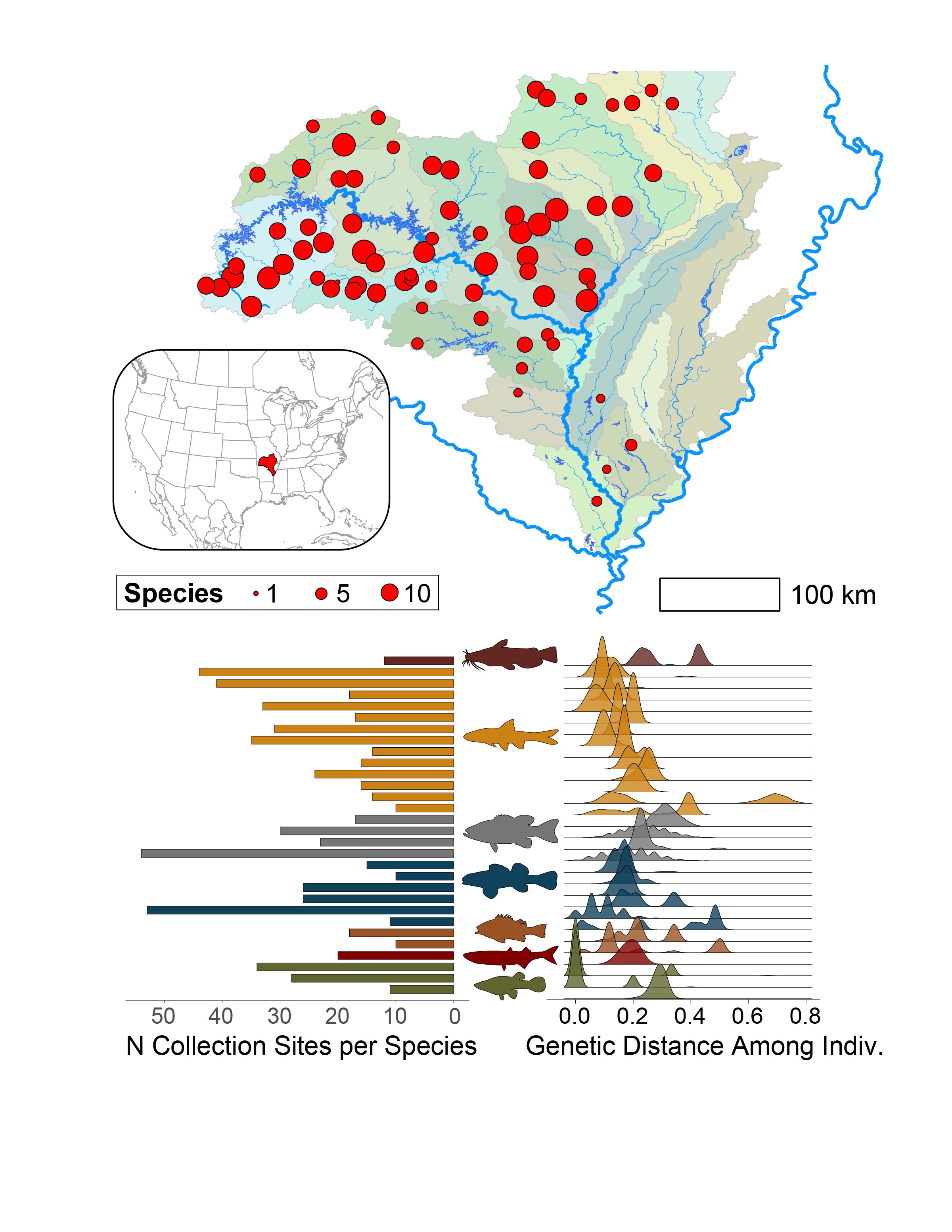
In collaboration with the Maryland Department of Natural Resources (DNR) and the Maryland Biological Stream Survey (MBSS), our lab uses eDNA to:
Check out my publications page for updates on eDNA research.
- Monitor biodiversity across diverse aquatic habitats throughout Maryland
- Track the presence and spread of rare, declining, or invasive species
- Compare eDNA detections to conventional sampling to evaluate monitoring efficiency
Check out my publications page for updates on eDNA research.